Constructing a Dynamic Model of "Actionable" Lung Cancer Subtypes
Cancers can be smart or dumb, and the smart ones are more difficult to deal with. Such is the case with lung cancer, which has a complex molecular heterogeneity. However, clearer insights into the molecular underpinnings of lung cancer and advances in diagnostics are enabling researchers to define the molecular subtypes of this cancer, aiding in the development of new therapeutic approaches.
In a paper published in the open-access journal PLoS ONE in February 2012, a multi-institutional and industry group defined “molecular subtypes” of lung cancer based on specific actionable genetic aberrations.1 The group had previously developed a formal process for classifying melanoma into molecular subtypes. The researchers defined a subtype as actionable if the mutation/aberration is associated with an approved assay and at least one FDA-approved or experimental targeted therapy with the potential for efficacy. For example, one subtype would be lung tumors with the EGFR exon 19 mutation, for which both commercial assays and targeted agents are available (the latest version of this model is available at http://bigmac.collabrx.com/lc_edit/index.php/File:Lung_SubtypeTable_1final.jpg).
Five oncogenes defining the subtypes with the current highest “strength of evidence” may be closest to arriving at the pharmacy as approved agents (Table).
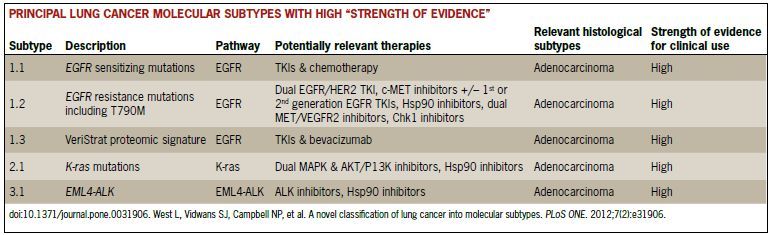
EGFR mutations are found in about 10% of Western populations and about 50% of Asians with lung adenocarcinoma. Data from several clinical trials show that progression-free survival is improved in patients treated with the EGFR inhibitors erlotinib or gefitinib over standard-of-care chemotherapy. In its 2011 updated guidelines for management of non-small cell lung cancer (NSCLC), the National Comprehensive Cancer Network (NCCN) called for EGFR testing in lung adenocarcinoma and large cell lung cancers upon recurrence of metastasis. In-progress or planned trials in recurrent or advanced NSCLC will determine the efficacy of approved and second-generation EGFR inhibitors in combination with agents such as MET/VEGFR2 inhibitors.
The T790 mutation is responsible for about 50% of cases of acquired resistance to erlotinib or gefinitib. Second-generation EGFR inhibitors such as afatinib (BIBW2992, SelleckBio), which irreversibly inhibits human EGFR2 (HER2) and EGFR kinases, are among potential therapeutic approaches to EGFR T790 mutations. MET inhibitors address the approximately 5% to 20% of patients with acquired resistance to EGFR inhibitors due to cMET amplification. These include adding crizotinib, foretinib, ARQ
197, and MetMAb to first-generation (eg, erlotinib) or second-generation EGFR tyrosine kinase inhibitors (TKIs). Heat shock protein 90 (Hsp90) inhibitors
(AUY922, ganetespib), which may block multiple signaling pathways that are not functioning properly in cancer cells, represent another approach to EGFR resistance mutations. Combination therapies such as a dual MET/VEGFR2 inhibitor, and others, are also under investigation.
The VeriStrat proteomic signature is a diagnostic tool for NSCLC patients who have tested negative for EGFR mutation. This lung cancer molecular subtype indicates likely responsiveness to EGFR inhibitor therapies such as erlotinib in the absence of EGFR mutations. Several studies have demonstrated that VeriStrat classification has significant predictive power. In one study, 73% of EGFR wild-type NSCLC patients who had previously received erlotinib and who had VeriStrat “good” status had improved survival after erlotinib treatment versus patients with VeriStrat “poor” status.
K-ras mutations have been reported in 15% to 20% of patients with NSCLC, and in about 30% to 50% of adenocarcinomas. Almost all patients with this mutation are smokers. This molecular subtype is associated with decreased response to EGFR TKIs. Potential therapeutic approaches to NSCLC patients within this group include MEK and mTOR inhibitors, Hsp90 inhibitors, and REOLYSIN (Reovirus Serotype 3-Dearing Strain, a proprietary formulation of human reovirus that lyses cancer cells, in particular those that have upregulated RAS signaling). REOLYSIN is currently in phase I, II, and III trials in NSCLC and other forms of cancer.
The EML4-ALK subtype is named for the EML4-ALK fusion oncogene, which produces a transforming tyrosine kinase with as many as nine different variants. Several isoforms are known to play a role in lung cancer. EML4-ALK is a novel target in a small subset of NSCLC patients who are typically younger women who are never to
light smokers. Therapeutic targets for this subtype of NSCLC include ALK inhibitors, such as the recently approved crizotinib (Xalkori, Pfizer) and LDK378 (Novartis). An Hsp90 inhibitor, IPI-504 (Infinity Pharmaceuticals), is in phase I and II trials. Ganetespib (Synta Pharmaceuticals) is also being studied as an ALK-targeted therapy.
In addition to these lung cancer molecular subtypes, which are accompanied by high strength of evidence, a number of secondary subtypes suggest that the landscape of future treatment options in NSCLC is promising.
Hoping to accelerate the process of translational research and to provide a conduit for open communication between basic research scientists, clinical trialists, and treating physicians, the authors of the PLoS ONE article designed an online lung cancer therapy finder, which can be found at www.collabrx.com/lung. Physicians who enter detailed information about a patient’s tumor can retrieve detailed current information about disease subtype, molecular testing options, suggestions for management of the tumor, and a list of current trials.
Reference
1. West L, Vidwans SJ, Campbell NP, et al. A novel classification of lung cancer into molecular subtypes. PLoS ONE. 2012;7(2):e31906.
